Tissue Processing for Stereology

By Krystal Sanchez and Peter R. Mouton
Modern stereology provides an unbiased (design-based) approach for quantifying stained biological objects, e.g., cells, fibers, capillaries, pathology deposits, tissue sections. These methods use stochastic geometry to avoid errors (bias) due to false assumptions, e.g., “assuming a cell is a sphere…” or models such as, e.g., “assuming fibers are projecting in all directions equally (isotropic)…” [1,2].
Beside avoiding false assumptions and incorrect models, reliable stereology data from biological specimens requires attention to proper tissue processing of specimens (e.g., mouse brain) to stained tissue sections for quantitative microscopic studies.
What is Tissue Processing For Stereology?
Tissue processing refers to the set of procedures for fixing, sectioning, and sampling tissue specimens for stereology analysis [3-5].
Fixation. Animal or human tissue are “fixed” to ensure tissue proteins and other cellular components remain as close to their in-vivo state as possible. Typical fixation for stereology studies uses aldehydes, e.g., 4% paraformaldehyde in a phosphate-buffered solution. These solutions of buffered aldehydes slowly penetrate the tissue, causing it to harden, which preserves and protects the tissue for subsequent tissue processing.
Dehydration. Besides fixing and preserving tissue, aldehydes are hydrophobic, i.e., they in effect drive water from the tissue, causing the tissue to shrink (dehydration).
Staining. Tissue processing includes treating tissue sections with dehydrating alcohols prior to staining. A typical alcohol series uses ethanol solutions at increasing concentrations, e.g., 60 to 100%. Like aldehydes, alcohols are hydrophobic, and therefore cause further tissue shrinkage until only tiny residues of water remain.
Clearing (for paraffin-embedding). Dehydration by fixatives and alcohol removes the structurally support from tissues. For paraffin (wax) embedding, a solvent (clearing agent) such as xylene that is immiscible, i.e., does not mix with alcohol, is used to infiltrate the tissue sample, thereby restoring stability. A clearing agent also adds transparency to the tissue, as well as removes fat deposits. As shown below, paraffin-embedding involves several more steps prior to cutting microtome sections.
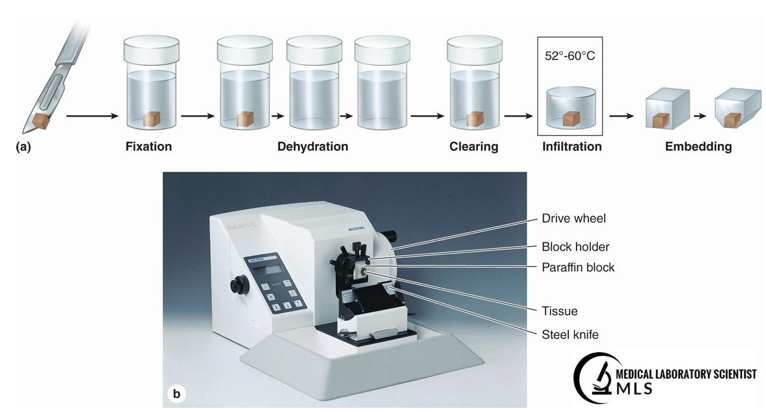
Figure 1: Tissue processing from fixation to embedding [3]
Stereological Tissue Processing Applications
Sections can be obtained by rotary microtome of paraffin-embedded sections (Figure 1), as frozen sections obtained using a sliding microtome or cryostat, or less frequently from a vibratome
Stereology Tissue Processing FAQ
The purpose of tissue processing is to generate high quality, well-stained tissue sections for microscopic analyses. To ensure proper tissue processing and sampling of tissue sections for stereology, it is highly recommended to use a reputable contract research organization (CRO) to process your tissue specimens.
The optimal amount of fixation combined with proper staining through the section thickness are two of the most critical steps.
This varies depending on the method. The low end of the range is histochemical staining, which requires a few hours to create frozen sections and stain a specimen with water-soluble stains such as hematoxylin & eosin (H&E) or cresyl violet. More complex methods are immunohistochemistry or immunofluorescence that can take up to several days for immunostaining techniques with primary and secondary antibodies.
Regardless of the staining method, a reliable stereology result requires ~ 8 to 10 tissue sections by systematic-uniform-random sampling (SURS) through an entire reference space (region of interest) [1-3, 7].
Professional tissue processing for stereology analyses typically costs ~ $300-500 per case.
Because of tissue shrinkage, stereology estimates on ex-vivo processed tissues will not reflect in-vivo parameters with one exception: counts of objects, e.g., cell counts, on properly processed tissue sections will estimate the true or expected values [1,2].
Tissue Sampling with Stereologer FAQ
High quality tissue sections allow for accurate and efficient quantification using the Stereologer system. After tissue processing is complete, further SURS sampling within each tissue section is done automatically by the Stereologer system. As a rule of thumb, one case requires about one hour for analysis using the Stereologer® system, provided the tissue sections are well-stained and properly sampled.
Once the microscopic field of views are automatically selected by SURS, stereology probes or test systems are automatically positioned over the sampling fields, and interactions between the object of interest and probes are counted either manually (user clicks); or automatically using a deep learning-based approach [6].
Typical stereology parameters include volume (3D), surface area (2D), length (1D) and Number (0D) and their variations.
The type of probe chosen depends on the dimensions of the geometric feature. For example, if a volume is the feature of interest, the probe chosen will be dimensionless, e.g., 0D points, using the Cavalieri principle [7]. For surface area, the 1D virtual cycloid probe is used [8]. If length of an object is required (e.g., capillaries, fibers), then a 2D probe (Space Balls®) is chosen [9]. If number is the parameter of interest, a 3D probe, the disector, is used in conjunction with the optical fractionator method [10]. The disector ensures that objects are sampled according to number rather than their size, shape, and orientation [1-3] while the optical fractionator provides an efficient and accurate estimate of total object number for the entire tissue of interest. The advantage of these parameter-probe combinations is that unbiased estimates will be obtained regardless of the tissue section orientation, e.g., coronal, sagittal, longitudinal.
Tissue sections in the range of 10 to 50 um thicknesses are usual for stereology studies by light microscopy, with thicker sections (>20 um) required for estimates using probes for Length [9], Surface area [8] and Number [10]. Stereology studies using electron microscopy and array tomography require ultra-thin sections in the nanometer range of thicknesses.
Stereology Analysis with the Stereologer system
At SRC Biosciences, our CRO services and technical staff have three decades of experience on advising clients on optimal tissue sampling procedures, sectioning intervals, staining protocols, and stereological test systems for hundreds of stereology projects.
They will advise you on:
- Tissue harvesting (removal), preservation, and shipping to our laboratories to ensure quality tissue section.
- Tissue processing for high quality sections as needed for your project.
- How to obtain the best possible results from the Stereologer software, a user-friendly program with automatic sampling and option for manual and automatic data collection using the correct probes and annotation tools [11].
Stereology Books, Manuals, and Journal Articles
[1] Mouton, P. R. (2002). Principles and Practices of Unbiased Stereology: An Introduction for Bioscientists (1st ed.). Johns Hopkins University Press.
[2] Mouton, P.R. (2011) A Concise Guide to Unbiased Stereology,” The Johns Hopkins University Press, Baltimore, MD.
[3] Brown, D. L. (2017). Practical Stereology Applications for the Pathologist. Veterinary Pathology, 54(3), 358–368. https://doi.org/10.1177/0300985817695781
[4] Geoffrey Rolls, B. A. S. (2019). An Introduction to Specimen Processing. Leica Biosystems. https://www.leicabiosystems.com/knowledge-pathway/an-introduction-to-specimen-processing/.
[5] Mahmoudzadeh Sagheb H R, Moudi B. Basic Application of Stereology in Histology and Medical Sciences, Gene Cell Tissue. 2014 ; 1(3):e24237. doi: 10.17795/gct-24237.
[6] Alahmari, S., Goldgof, D., Hall, D. Phoulady, H.A. Patel, R., Mouton, P.R. Automated Counts of Stained Cells by Deep Learning and Unbiased Stereology. J Chem Neuroanat, 96:94-101, 2019.
[7] Gundersen, H.J.G.; Jensen, E.B.; Kiêu, K.; Nielsen, J. (1999) The efficiency of systematic sampling in stereology–reconsidered. J Microsc. 193(3), 199-211.
[8] Gokhale AM, Evans RE, Mackes JL, Mouton PR. Stereological Estimation Of Surface Area In Thick Transparent Sections Of Arbitrary Orientation Using Virtual Cycloids. J Microsc 216(Pt1): 25-31, 2004.
[9] Mouton PR, Gokhale AM, Ward NL, West MJ. Stereological Length Estimation Using Spherical Probes. J Microsc 206: 54-64, 2002.
[10] West, M.J.; Slomianka, L.; Gundersen, H.J.G. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 231(4), 482-497, 1991.
[11] Mouton, P.R. The Stereologer Handbook: Introduction to Unbiased Stereology and Computerized Stereology (2011), 339 pp, SRC Publications, Tampa, Florida.
2024 Cade Prize Finalist
PainPredict: AI-based Early Pain Detection for Avoidance of Neonatal Pain in the NICU. PainPredict™, a new invention from the AI team at SRC Biosciences and
Tissue Processing for Stereology
By Krystal Sanchez and Peter R. Mouton Modern stereology provides an unbiased (design-based) approach for quantifying stained biological objects, e.g., cells, fibers, capillaries, pathology deposits,
SRC Biosciences Receives National Science Foundation (NSF) Technology Enhancement for Commercial Partnerships (TECP) Award
In May 2021 the National Science Foundation (NSF) awarded a Technology Enhancement for Commercial Partnerships (TECP) Award to the Stereology Resource Center (SRC Biosciences® or