Whole Slide Imaging for Unbiased Stereology
What Is Whole Slide Imaging?
Whole slide imaging (WSI) is the digitization of tissue sections on glass slides. The tissue section rendered locally on the computer by a WSI software can be viewed remotely using the WSI software, navigated using a simulated slide viewer interface, and analyzed with tissue analysis tools.
In combination with unbiased stereology software, WSI allows for seamless data collection and analysis using state-of-the-art methods.
Digitalization of pathology tissues, advances in analysis and technology such as cloud storage options, computing processing power, and fast data transfer speeds have facilitated the popularity of WSI software.
WSI software has been used for archiving tissue images, virtual microscope web-viewers allowing for the emulation of a light microscope at different magnifications, and tissue analysis for stereology. [1]
Whole Slide Image Acquisition
Methods of image acquisition include using digital whole slide imaging scanners or fundamental computerized microscope equipped with motorized XY or XYZ stage control as appropriate.
In the first step, digitizing hardware converts the tissue image into a digital image file, typically at 40x objective power or below.
Tissue sections mounted on glass slides are scanned in a variety of orientations either manually or automatically.
- Tile-based scanning: A robotic controlled slide stage holds many image frames and assembles them in a mosaic image with proper autocorrelation to ensure proper alignment. [1]
- Line-based scanning: A servomotor-based slide stage produces a group of images in a single axis of acquisition. [1]
- Brightfield: Most common and cost-effective option for stained tissue section [2].
- Fluorescence: Used to digitalized fluorescently stained slides such as in immunohistochemistry [2].
- Multispectral: Captures spectral information from a slide can be adapted to brightfield and fluorescence applications. [2]
Methods of slide acquisition differ in not only the process by which the image is captured but the specifications of the resultant image. Scanning at higher magnifications, digitization of larger tissue sections, and digitization of higher contrast images like those of fluorescence lead to longer slide acquisition times and a requirement for larger storage spaces.
Furthermore, it is important to select an adequate image resolution. While the slide can be viewed under any objective lens, enlarging an image beyond the true resolution will lead to pixelization and will cause the image to look out of focus. How a scan looks is dependent on the size and number of pixels the computer monitor can support. Thus, if the camera sensor has a lower resolution than the objective’s numerical aperture, information will be lost, and even the use of a high-resolution monitor will not increase resolution when displaying a scan produced by this system. [2] To reproduce magnifications necessary for clinical and research applications, the correct resolution and color depth should be used.
Whole Slide Analysis
The second process of WSI involves specialized WSI software reading these image files and allowing the researcher to perform tissue analysis, such as a researcher annotating tissue sections in a z-stack in a stereology application.
WSI software can be locally based or web based. This specialized software for WSI enables users to perform fundamental functions of light microscopy such as panning and zooming around a virtual slide at different magnifications. Other analysis tools allow for viewing the slide at unconventional magnifications, annotation tools such as measurements and shape overlays, or sophisticated design-based analysis methods such as stereology. [3]
Measurements taken during whole slide analysis include area-based assessments to quantify 2D areas in a chemical or an immunohistochemical stain such as fat or cell-based to quantify pathologically-affected cells in neurodegenerative diseases.
Several use cases for subsequent whole slide analysis are further explained below.
- Slide Archival: With advancements in cloud storage, the cumbersome task of maintaining physical slides prone to human and environmental degradation will be eliminated. Creating a virtual slide maintains the image quality and allows for future analysis after years of storage. [3]
- Telepathology: In core research facilities, data collectors must perform tissue analysis if the lab does not have required microscopes. Thus, the ability to digitally store tissue slides allows for remote tissue analysis. [3]
- Education: Accessibility and convenience to WSI educational material allows for more efficient teaching and standardization of analysis to maintain a high level of competency. [3]
- Research: Histological applications by pharmaceutical and biotechnology companies include automated histology (tissue storage, sectioning, staining), slide scanning, and image management. WSI enables researchers to remotely view stains, compare multiple slides at a time, and archive slides before they are sacrificed to use their cellular material. [3]
Whole Slide Imaging Scanners
Microscope Slide Scanners
The essential components of a Whole Slide Image Scanner include [4]:
- Microscope with required lens objectives
- Light source (brightfield/fluorescence)
- Motorized stage to move the slides around, select new slides, etc.
- Digital cameras for image capture
- Computer with specified processing power and corresponding monitor
- Specialized software to capture, analyze, store tissue images
Stereology on Whole Slide Images
Stereology uses unbiased sampling and estimation methods to quantify tissue with first-order stereological parameters such as volume (V), surface area (S), length (L), and number (N). The integration of stereology and whole slide imaging offers a design-based approach for analysis of these digital images.
Software for Whole Slide Analysis
General stereology software requirements include the ability to set study parameters, acquire tissue images, perform data collection, and output study results. General parameters to be set are XYZ step size, number of sections, section sampling interval, etc. Based on these parameters, images will be captured within a set tissue thickness. These images will be analyzed by trained data collectors using stereology probes; each company will have their own probe designs.
The Stereologer automatically determines optimal study parameters, acquires tissue images, guides data collection and outputs study results to saved data files. The user can either specify the storage of the images locally on the computer or select a cloud or external hard drive. Below is the whole slide imaging method available to users using SRC Biosciences’ stereology software.
Digital Stereology™ refers to the automatic unbiased sampling and manual analysis of z-axis stacks of digital images, i.e., disector stacks. Digital images provide the data for analyses with the ability to archive permanent evidence of collected data.
There are two options for stereological analysis of digital images:
- “Volume-based” manual counts. The user scrolls through each z-stack (disector stack) in real-time using the up/down arrows while counting (clicking) on the stained objects of interest.
- Digital Videos. Each 3D stack of z-axis images is automatically converted into a digital video and presented for rapid data collection at speeds controlled by the user.
Hardware for Whole Slide Analysis
Microscope
Microscopes in stereology systems allow for stage movement in the XYZ positions, tracking of cell location, and speed modulation during analysis. Stereologer is compatible with all microscopes, for example, Zeiss, Leica, Olympus, and Nikon. Many modern microscopes include an internally controlled motorized XYZ stage control. If a microscope is missing this required hardware, we add them as external devices from reliable companies such as Applied Scientific Instrumentation, Prior Scientific and Ludl.
Camera
During the initial inquiry stage, hardware specialists determine if a client’s current camera system is compatible with the stereology software. Companies that specialize in stereology systems maintain relationships with many camera vendors, such as Zeiss, Nikon, Olympus, and Leica, to determine the best camera for a lab’s research goals and budgetary limits. For whole slide imaging, resolution is also considered. All third-party cameras follow industry standards, such as DCAM and USB3.
Computer
All new PC computers support the Windows 10 OS and include PCIe boards for plug-in support of cameras and motorized stage connections. Preferred configurations for Stereologer systems include Intel Core i7 processor and memory of 16 GB or higher. As the needs for processing images increase with whole slide imaging, preferred HD storage is minimum 500 GB with Solid State Drives (SSD).
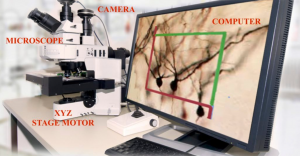
Whole Slide Imaging with Stereologer
Stereologer offers software capabilities enabling users to perform whole slide imaging, allowing for remote tissue analysis and intuitive collaboration. The determination of whether your microscope system meets our software requirements will be made by our Stereologer support team.
Works Cited
[1] Beckwith, B. A. (2016). Standards for Digital Pathology and Whole Slide Imaging. Digital Pathology, 87–97. https://doi.org/10.1007/978-3-319-20379-9_9
[2] Farahani N, Parwani A, Pantanowitz L. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathology and Laboratory Medicine International. 2015;7:23-33 https://doi.org/10.2147/PLMI.S59826
[3] Fathima S, Parwani A. WSI fundamentals. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/informaticswholeslidefund.html. Accessed May 24th, 2021.
[4] Mark D. Zarella, Douglas Bowman;, Famke Aeffner, Navid Farahani, Albert Xthona;, Syeda Fatima Absar, Anil Parwani, Marilyn Bui, Douglas J. Hartman; A Practical Guide to Whole Slide Imaging: A White Paper From the Digital Pathology Association. Arch Pathol Lab Med 1 February 2019; 143 (2): 222–234. doi: https://doi.org/10.5858/arpa.2018-0343-RA

